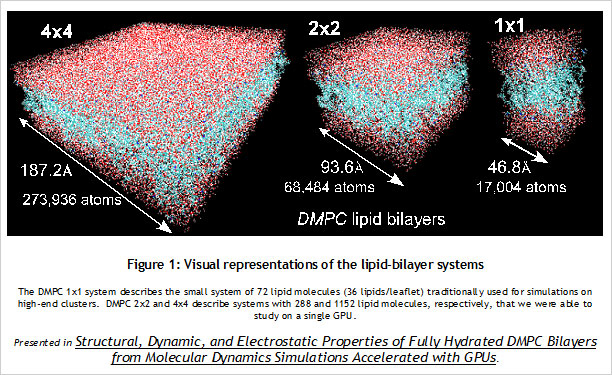
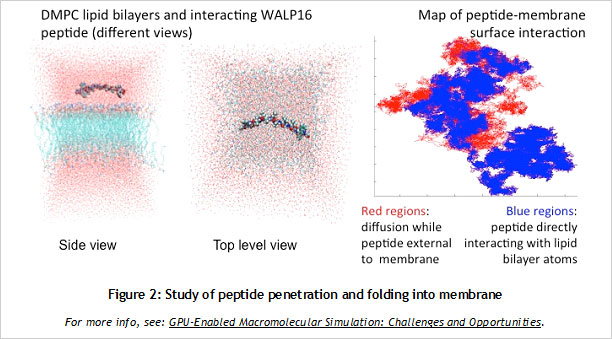
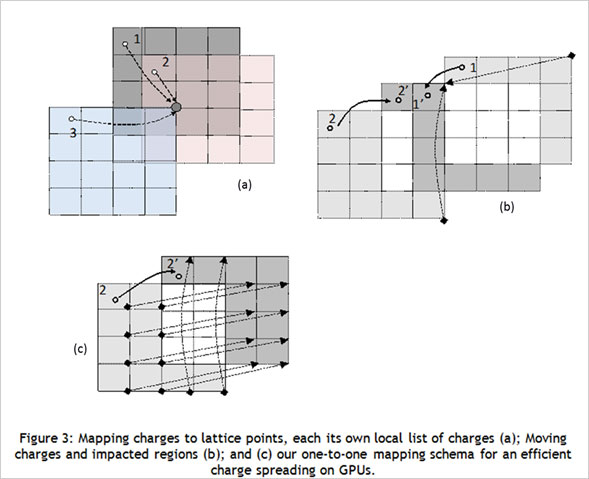
CUDA Spotlight: Michela TauferBy Calisa Cole, posted August 21, 2014 GPU-Accelerated Scientific ComputingThis week's Spotlight is on Dr. Michela Taufer, Associate Professor at the University of Delaware. Michela heads the Global Computing Lab (GCLab), which focuses on high performance computing (HPC) and its application to the sciences. This interview is part of the CUDA Spotlight Series.Q & A with Michela TauferNVIDIA: Michela, what is the mission of the Global Computing Lab at the University of Delaware? Interdisciplinary research with scientists and engineers in fields such as chemistry and chemical engineering, pharmaceutical sciences, seismology, and mathematics is at the core of our activities and philosophy. NVIDIA: Tell us about your work with GPUs. NVIDIA: Can you provide an example? With our code empowered with the PME components, we could move the traditional scale for studying membranes like DMPC lipid bilayers from membranes on the order of 72 lipid molecules (17,004 atoms) to 16-times larger membranes of 1,152 lipid molecules (27,3936 atoms) in explicit solvent [see Figure 1]. NVIDIA: How do GPUs help you in your work today? I will never forget the excitement of my collaborator Sandeep Patel when we were able to observe the exploration activity of small peptides on a large membrane surface. It was known that the phenomenon existed but it was not observable previously because of the complexity of simulating membranes at large space and time scales. But we did it! We could clearly identify the location on the membrane when the peptide was external to the membrane (red dots on the peptide-membrane surface in Figure 2) versus when the peptide was directly interacting with the lipid bilayer atoms (blue dots on the peptide-membrane surface in Figure 2). This would have not been possible without GPUs, which provided us with the computing power to cope with the large length and time scales for this type of molecular simulation. NVIDIA: What approaches have you used to apply the CUDA platform to your work? In this step of the MD simulation, charges are spread around a neighborhood of 4x4x4 lattice points and multiple charges can contribute to the total charge at each point. The process involves placing the charges on lattice points and accumulating the impact of charges for each point and simulations step (as shown in Figure 3.a where the impact of three charges sums up in one lattice point). A local list of charges can be built for each lattice point. The critical step in the computation is to efficiently update this list as the particles move.
When a charge moves due to dynamics of the system (as shown in Figure 3.b where a charge moves from Point 2 to Point 2'), we can identify three regions of the lattice: The lattice points gaining the charge impact are initially not aware of the charge movement and thus need to search through the global list of charges for the information when an update takes place. This search is the killing point of the performance but it was also where the understanding of the physical process made the difference for us. We observed that the movement of a charge produces an equal number of lattice points gaining the charge and points losing the charge. Lattice points losing a charge are aware of the destination points in a one-to-one mapping schema shown in Figure 3.c. This was our "Aha!" moment! We immediately thought we could benefit from this one-to-one mapping to reduce the time for the update of the lists by having the threads of the points losing the charge updating the lists of the points gaining the charge. This prevents the lattice points gaining the charge impact from having to explore the whole global list of charges and made the difference in the PME performance when executed completely on GPU [See: FEN ZI: GPU Enabled Molecular Dynamics Simulation of Large Membrane Regions Based on CHARMM Force Field and PME]. NVIDIA: Which CUDA resources do you recommend? NVIDIA: What hardware and software are you using? NVIDIA: How did you first learn about CUDA? I had read about the Brook language and was considering writing my MD code in that language. Marc changed my mind. While speaking with him I immediately saw the potential hidden in that small memory stick. I returned to my university and started working with one of my students on our MD code for GPUs. In 2008 we finalized the first version of our code; in April 2009 we published our work on what would become the FEN ZI MD code. Today FEN ZI is available for the public as open-source software and it has been fundamental for me and my colleague Sandeep Patel in the study of membranes and membrane penetrations. NVIDIA: What excites you the most about your work? I feel I have a very special relationship with GPUs because they helped me to reveal my “MacGyver spirit”. At the very beginning of the GPU era, the few tools and libraries that were available at the time were incomplete and I had to rely on my creativity and imagination to rethink algorithms and applications. GPUs empowered me to build an “explosive” solution to my algorithmic problems using a Swiss army knife and bubble gum. Today, of course, there is a robust ecosystem of tools, libraries, applications, companies, research labs and other resources built around GPU computing, making it much more accessible. I am happy to be a member of the select group of people who have attended every single GPU Tech Conference from the very beginning, starting with the NVISION 2008 meeting in which we (the HPC people) were relegated in a small corner of the convention center in San Jose. Before leaving for NVISION I remember I spoke with a colleague in graphics who was telling me how HPC on GPUs was "hopeless." This year at GTC 2014, I was looking around at all the innovation and breakthroughs, and thinking back to the 2008 meeting, where a small group of HPC people had a big vision and lots of courage. I am pleased my colleague was wrong and am so proud to be part of this very special community. Bio for Michela TauferMichela Taufer joined the University of Delaware in 2007, where she was promoted to associate professor with tenure in 2012. She earned her M.S. degree in Computer Engineering from the University of Padova and her Ph.D. in Computer Science from the Swiss Federal Institute of Technology (ETH). She was a post-doctoral research supported by the La Jolla Interfaces in Science Training Program (also called LJIS) at UC San Diego and The Scripps Research Institute. Before she joined the University of Delaware, Michela was faculty in Computer Science at the University of Texas at El Paso. Michela has a long history of interdisciplinary work with high-profile computational biophysics groups in several research and academic institutions. Her research interests include software applications and their advanced programmability in heterogeneous computing (i.e., multi-core platforms and GPUs); cloud computing and volunteer computing; and performance analysis, modeling and optimization of multi-scale applications. She has been serving as the principal investigator of several NSF collaborative projects. She also has significant experience in mentoring a diverse population of students on interdisciplinary research. Michela's training expertise includes efforts to spread high-performance computing participation in undergraduate education and research as well as efforts to increase the interest and participation of diverse populations in interdisciplinary studies. Michela has served on numerous IEEE program committees (SC and IPDPS among others) and has reviewed for most of the leading journals in parallel computing.Relevant Links http://www.cis.udel.edu/~tauferhttp://gcl.cis.udel.edu Active Projects and Software Repositories Docking@Home: http://docking.cis.udel.edu ExSciTecH: https://exscitech.org/ QCNExplorer: http://qcnexplorer.org/ GitHub: https://github.com/TauferLab Follow Michela on Twitter at: https://twitter.com/MichelaTaufer Follow Michela's Group on Twitter at: https://twitter.com/TauferLab Contact Info Michela Taufer David and Beverly J.C. Mills Career Development Chair Computer and Information Sciences, Biomedical Engineering University of Delaware, 101 Smith Hall, Newark, DE 19716 E-Mail: taufer@acm (dot) org # # # |