Get started today with this GPU Ready Apps Guide.

AMBER is a molecular dynamics application developed for the simulation of biomolecular systems. AMBER is GPU-accelerated and runs both explicit solvent PME and implicit solvent GB simulations.
The latest version, AMBER 16, runs up to 15X faster on NVIDIA GPUs over CPU-only systems*, enabling users to run biomolecular simulations in hours instead of days. AMBER is distributed under a license from the University of California.
*Dual-CPU server: Intel E5-2690 v4 @ 2.6GHz | 3.5GHz Turbo (Broadwell-EP) | NVIDIA® Tesla® P100 | ECC off | Autoboost ON | PME-JAC_NVE_2fs dataset
Installation
System Requirements
AMBER is portable to any version of Linux with C, C++ and Fortran 95 compilers. AMBER is very GPU-intensive, so the latest GPUs will deliver the highest performance. See the recommended system Configurations section for more details.
Download and Installation Instructions
AMBER is distributed in source code form by purchasing a license. Details can be found on the application website.
BELOW IS A BRIEF SUMMARY OF THE COMPILATION PROCEDURE
1. Download the AMBER and ambertools source code archives
2. Install dependencies. For information on installing AMBER’s dependencies on a variety of Linux distributions click here. For more information, see the official reference manual.
3. Extract the packages and set the AMBERHOME environment variable (shown here for bash)
tar xvjf AmberTools16.tar.bz2
tar xvjf Amber16.tar.bz2
cd amber16
export AMBERHOME=`pwd` # bash, zsh, ksh, etc.
setenv AMBERHOME `pwd` # csh, tcsh
4. Configure AMBER
The build script may need to update AMBER and/or ambertools. Select ‘y’ each time to accept when prompted.
./configure -cuda -noX11 gnu # With CUDA support, GNU compilers
./configure -cuda -mpi -noX11 gnu # With CUDA and MPI support, GNU compilers
5. Build AMBER
make install -j8 # use 8 threads to compile
If you wish to run both single and multiple-GPU runs, you must perform the configure and build steps for both with MPI and without.
./configure -cuda -noX11 gnu
make install -j32
./configure -cuda -mpi -noX11 gnu
make install -j32
To perform CPU-only benchmarking runs, you also need to build a non-CUDA-enabled binary.
./configure -mpi -noX11 gnu
make install -j32
Running Jobs
Before running a GPU-accelerated version of AMBER, install the latest NVIDIA display driver for your GPU.
To run AMBER, you need the ‘pmemd.cuda’ executable. For multi-GPU runs you also need the ‘pmemd.cuda.MPI’ executable (see download and installation instructions for compiling with MPI support). For multi GPU runs, GPUs should be in the same physical node and the GPUs used need to support peer to peer communication to get the best possible performance.
You should also download the AMBER16 benchmark suite, freely available from their benchmark page. From this point we will assume you have downloaded the benchmarks and extracted them to a directory we will refer to as $BENCHMARKS.
COMMAND LINE OPTIONS
It is recommended to set your GPU boost clocks to maximum when running AMBER in order to obtain best performance. In this case we set the clocks for GPU 0, using the boost clocks for a Tesla K80 on the first line, a Tesla M40 on the second, and a Tesla P100 on the third. Note that Geforce devices may not have modifiable boost clocks.
nvidia-smi -i 0 -ac 2505,875 # K80
nvidia-smi -i 0 -ac 3004,1114 # M40
Nvidia-smi -i 0 -ac 715,1328 # P100
General command line to run AMBER on a single-node system:
$AMBERHOME/bin/pmemd.cuda -i {input} -o {output file} {options}
On a multi-GPU system AMBER has to be run with mpirun as specified below:
mpirun -np <NP> $AMBERHOME/bin/pmemd.cuda.MPI -i {input} -o {output} {options}
{options}
1. -O : Overwrite the output file if it already exists.
2. -o : specify the filename for output to be written to
It is recommended to always check the beginning of the output file to make sure that all 87 parameters, in particular the GPUs have been specified correctly.
{input}
Use corresponding input file from one of the datasets in the next sub-section (generally mdin.GPU)
Example 1. Run JAC production NVE (DHFR (NVE) 2fs from the benchmarks page) on a single GPU:
cd $BENCHMARKS/PME/JAC_production_NVE
$AMBERHOME/bin/pmemd.cuda -i mdin.GPU -O -o mdout
Example 2. Run Cellulose production NPT on 2 GPUs (all on the same node):
cd $BENCHMARKS/PME/Cellulose_production_NPT
mpirun -np 2 $AMBERHOME/bin/pmemd.cuda.MPI -i mdin.GPU -O -o mdout
Example 3. Run Cellulose production NPT on 32 CPU cores (all on the same node):
cd $BENCHMARKS/PME/Cellulose_production_NPT
mpirun -np 32 $AMBERHOME/bin/pmemd.MPI -i mdin.CPU -O -o mdout
Benchmarks
This section demonstrates GPU acceleration for selected datasets. The benchmarks are listed in increasing number of atoms order. When reading the output the figure of merit is "ns/day" (the higher the better), located at the end of the output in the "mdout" file. It is best to take the measurement over all time steps (instead of the last 1000 steps).

# of Atoms
23,558
Typical Production MD NPT with GOOD energy conservation, 2fs.
&cntrl
ntx=5, irest=1,
ntc=2, ntf=2, tol=0.000001,
nstlim=250000,
ntpr=2500, ntwx=2500,
ntwr=250000,
dt=0.002, cut=8.,
ntt=0, ntb=1, ntp=0,
ioutfm=1,
/
&ewald
dsum_tol=0.000001,
/
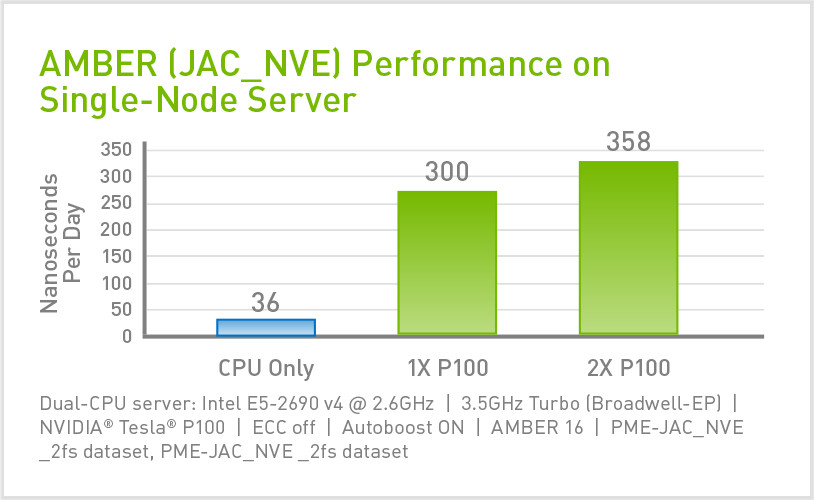
DHFR (JAC PRODUCTION) NPT 2FS
JAC (Joint Amber CHARMM) Production NPT is DHFR in a TIP3P water box. It is one of the smaller benchmarks with 23,588 atoms including water. The benchmark is run with an explicit solvent model (PME).

# of Atoms
90,906
Typical Production MD NPT with GOOD energy conservation.
&cntrl
ntx=5, irest=1,
ntc=2, ntf=2, tol=0.000001,
nstlim=70000,
ntpr=1000, ntwx=1000,
ntwr=10000,
dt=0.002, cut=8.,
ntt=0, ntb=1, ntp=0,
ioutfm=1,
/
&ewald
dsum_tol=0.000001
/
FACTOR IX NPT
FactorIX is a benchmark that simulates the blood coagulation factor IX. It consists of 90,906 atoms including water. Water is modelled as an explicit TIP3P solvent. This benchmark is run with an explicit solvent model (PME).

# of Atoms
408,609
Typical Production MD NVT
& cntrl
ntx=5, irest=1,
ntc=2, ntf=2,
nstlim=14000,
ntpr=1000, ntwx=1000,
ntwr=8000,
dt=0.002, cut=8.,
ntt=1, tautp=10.0,
temp0=300.0,
ntb=2, ntp=1, barostat=2,
ioutfm=1,
/
CELLULOSE NPT
This benchmark simulates a cellulose fiber. It is one of the larger models to be simulated in the benchmark with 408,609 atoms including water. Water is modelled as an explicit TIP3P solver. The benchmark is run with an explicit solvent model (PME).
Expected Performance Results
This section provides expected performance benchmarks for different across single and multi-node systems.


Recommended System Configurations
Hardware Configuration
PC
Parameter
Specs
CPU Architecture
x86
System Memory
16-32GB
CPUs
1-2
GPU Model
NVIDIA TITAN X
GPUs
1-2
Servers
Parameter
Specs
CPU Architecture
x86
System Memory
64-128GB
CPUs
1-2
GPU Model (for workstations)
Tesla
® P100
GPU Model (for servers)
Tesla
® P100
GPUs
1-2 P100 PCIe
1-4 P100 SMX2
1-4 P100 SMX2
Software Configuration
Software stack
Parameter
Version
Application
AMBER 16
OS
Centos 7.2
GPU Driver
375.20 or newer
CUDA Toolkit
8.0 or newer
See a list of supported GPUs on Amber.
