Get started today with this GPU Ready Apps Guide.

NAMD (NAnoscale Molecular Dynamics) is a production-quality molecular dynamics application designed for high-performance simulation of large biomolecular systems. Developed by University of Illinois at Urbana-Champaign (UIUC), NAMD is distributed free of charge with binaries and source code.
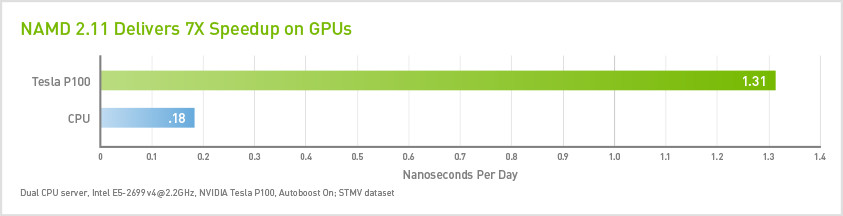
The latest version, NAMD 2.11, typically runs 7x faster on NVIDIA GPUs over CPU-only systems*, enabling users to run molecular dynamics simulations in hours instead of days. It’s also up to 2x faster than NAMD 2.10, which helps users save on hardware cost while significantly accelerating their scientific discoveries.
*Dual CPU server, Intel E5-2698 v3@2.3GHz, NVIDIA Tesla K80 with ECC off, Autoboost On; STMV dataset
Installation
System Requirements
NAMD should be portable to any parallel platform with a modern C++ compiler. Precompiled GPU-accelerated NAMD 2.11 binaries are available for download for Windows, Mac OS X, and Linux. NAMD makes use of both GPU and CPU, therefore we recommend to have a relatively modern CPU to achieve the best NAMD application performance.
Download and Installation Instructions
NAMD developers provide complete, optimized binaries for all common platforms to which NAMD has been ported. The binaries can be obtained from the NAMD download page.
It should not be necessary to compile NAMD unless you want to add or modify features, improve performance by building a Charm++ version that takes advantage of special networking hardware, or if you have customized your version of NAMD. Complete directions for compiling NAMD are contained in the release notes, and are available on the NAMD web site and are included in all distributions.
BELOW IS A BRIEF SUMMARY OF THE COMPILATION PROCEDURE
1. Download NAMD Source code.
2. Extract the package and Charm++
tar xf NAMD_2.11_Source.tar.gz
cd NAMD_2.11_Source
tar xf charm-6.7.0.tar
3. Download and install TCL and FFTW.
Please see the release notes for the appropriate version of these libraries.
wget http://www.ks.uiuc.edu/Research/namd/libraries/fftw-linux-x86_64.tar.gz
wget http://www.ks.uiuc.edu/Research/namd/libraries/tcl8.5.9-linux-x86_64.tar.gz
wget http://www.ks.uiuc.edu/Research/namd/libraries/tcl8.5.9-linux-x86_64-threaded.tar.gz
tar xzf fftw-linux-x86_64.tar.gz
mv linux-x86_64 fftw
tar xzf tcl*-linux-x86_64.tar.gz
tar xzf tcl*-linux-x86_64-threaded.tar.gz
mv tcl*-linux-x86_64 tcl
mv tcl*-linux-x86_64-threaded tcl-threaded
4. Build Charm++
The build script has an interactive mode, you can also specify options in the command line as explained below.
cd charm-6.7.0
./build charm++ {arch} {C compiler} {Fortran compiler} {other options}
For {arch}, it is required to use one of the following options: 'verbs-linux-x86_64' for multiple node configurations, 'multicore-linux64' for a single node system. Other options are not required but recommended for best performance: 'smp' to use SMP for multi-node builds (Click here for more details), '--with-production' to use production level optimizations.
Example 1. Charm++ build for multi-node configuration using Intel compilers and SMP with all production optimizations:
./build charm++ verbs-linux-x86_64 icc smp --with-production
Example 2. Charm++ build for single-node configuration using Intel compilers with all production optimizations.
./build charm++ multicore-linux64 icc --with-production
5. Build NAMD.
Make sure you're in the main NAMD directory, then configure and run make:
./config {namd-arch} --charm-arch {charm-arch} {opts}
cd {namd-arch}
make -j4
For {namd-arch} we recommend 'Linux-x86_64-icc', {charm-arch} should be the same arch that you built Charm++ with, {opts} should be set to '--with-cuda' to enable GPU acceleration, otherwise set to '--without-cuda' for CPU-only build. The build directory can be extended with .[comment] to build, e.g., Linux-x86_64.cuda. This is helpful to manage different builds of NAMD. Note that the extension does not have any meaning, so './config Linux-x86_64.cuda' won't be a CUDA build unless you also specify '--with-cuda'.
Optional flags for the CUDA build:
a. --cuda-prefix can be used to set correct path to your CUDA toolkit;
b. --cuda-gencode allows you to add different GPU compute architecture.
Example 1. NAMD build for x86_64 architecture using Intel compilers, with GPU acceleration and specified CUDA path, for single node:
./config Linux-x86_64-icc --charm-arch multicore-linux64-icc --with-cuda --cuda-prefix /usr/local/cuda
Example 2. NAMD build for x86_64 architecture using Intel compilers, with GPU acceleration and specified CUDA path, for multiple nodes:
./config Linux-x86_64-icc --charm-arch verbs-linux-x86_64-smp-icc --with-cuda --cuda-prefix /usr/local/cuda
Example 3. NAMD build for x86_64 architecture using Intel compilers, without GPU acceleration, for multiple nodes:
./config Linux-x86_64-icc --charm-arch verbs-linux-x86_64-smp-icc
Running Jobs
Before running a GPU-accelerated version of NAMD, install the latest NVIDIA display driver for your GPU. To run NAMD, you need ‘namd2’ executable and for multi-node run you also need the charmrun executable. Make sure to specify CPU affinity options as explained below.
COMMAND LINE OPTIONS
General command line to run NAMD on a single-node system:
namd2 {namdOpts} {inputFile}
On a multi-node system NAMD has to be run with charmrun as specified below:
charmrun {charmOpts} namd2 {namdOpts} {inputFile}
{charmOpts}
- ++nodelist {nodeListFile} - multi-node runs require a list of nodes
- Charm++ also supports an alternative ++mpiexec option if you're using a queueing system that mpiexec is setup to recognize.
- ++p $totalPes - specifies the total number of PE threads
- This is the total number of Worker Threads (aka PE threads). We recommend this to be equal to (#TotalCPUCores - #TotalGPUs).
- ++ppn $pesPerProcess - number of PEs per process
- We recommend to set this to #ofCoresPerNode/#ofGPUsPerNode – 1
- This is necessary to free one of the threads per process for communication. Make sure to specify +commap below.
- Total number of processes is equal to $totalPes/$pesPerProcess
- When using the recommended value for this option, each process will use a single GPU
{namdOpts}
- NAMD will inherit '++p' and '++ppn' as '+p' and '+ppn' if set in {charmOpts}
- Otherwise, for the multi-core build use '+p' to set to the number of cores.
- It is recommended to have no more than one process per GPU in the multi-node run. To get more communication threads, it is recommended to launch exactly one process per GPU. For single-node it is fine to use multiple GPUs per process.
- CPU affinity options (see user guide):
- '+setcpuaffinity' in order to keep threads from moving about
- '+pemap #-#' - this maps computational threads to CPU cores
- '+commap #-#' - this sets range for communication threads
- Example for dual-socket configuration with 16 cores per socket:
- +setcpuaffinity +pemap 1-15,17-31 +commap 0,16
- GPU options (see user guide):
- '+devices {CUDA IDs}' - optionally specify device IDs to use in NAMD
- If devices are not in socket order it might be useful to set this option to ensure that sockets use their directly-attached GPUs, for example, '+devices 2,3,0,1'
We recommend to always check the startup messages in NAMD to make sure the options are set correctly. Additionally, ++verbose option can provide a more detailed output for the execution for runs that use charmrun. Running top or other system tools can help you make sure you’re getting the requested thread mapping.
./namd2 +p 20 +devices 0,1 apoa1.namd
Example 2. Run STMV on 2 nodes, each node with 2xGPU and 2xCPU (20 cores) and SMP NAMD build (note that we launch 4 processes, each controlling 1 GPU):
charmrun ++p 36 ++ppn 9 ./namd2 ++nodelist $NAMD_NODELIST +setcpuaffinity +pemap 1-9,11-19 +commap 0,10 +devices 0,1 stmv.namd
Note that by default, the "rsh" command is used to start namd2 on each node specified in the nodelist file. You can change this via the CONV_RSH environment variable, i.e., to use ssh instead of rsh run "export CONV_RSH=ssh" (see NAMD release notes for details).
Benchmarks
This section demonstrates GPU acceleration for selected datasets. Note that you may need to update the input files to reduce the output energy/timing frequency. The benchmarks are listed in increasing number of atoms order. It is recommended to run large test cases like STMV on systems with multiple GPUs.

# of Atoms
92,224
Time Steps
500
Output Energies
100
Output Timing
100
APOA1
Apolipoprotein A1 (ApoA1) is the major protein component of high-density lipoprotein (HDL) in the bloodstream and plays a specific role in lipid metabolism. The ApoA1 benchmark consists of 92,224 atoms and has been a standard NAMD cross-platform benchmark for years.

# of Atoms
327,506
Time Steps
500
Output Energies
100
Output Timing
100
ATPASE
ATP synthase is an enzyme that synthesizes adenosine triphosphate (ATP), the common molecular energy unit in cells. The F1-ATPase benchmark is a model of the F1 subunit of ATP synthase.

# of Atoms
1,066,628
Time Steps
500
Output Energies
100
Output Timing
100
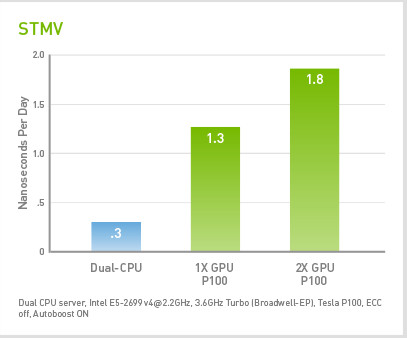
STMV
Satellite Tobacco Mosaic Virus (STMV ) is a small, icosahedral plant virus that worsens the symptoms of infection by Tobacco Mosaic Virus (TMV). SMTV is an excellent candidate for research in molecular dynamics because it is relatively small for a virus and is on the medium to high end of what is feasible to simulate using traditional molecular dynamics in a workstation or a small server.
READING THE OUTPUT
The figure of merit is "ns/day" (the higher the better) but is printed in the log as "days/ns" (the lower the better). Note that most molecular dynamics users focus on “ns/day”, the inverted value of “days/ns”. It is recommended to take the average of all occurrences of this value for benchmarking purposes; the frequency of the output can be controlled in the corresponding *.namd input file. Due to initial load balancing the "Initial time:" outputs should be neglected in favor of the later "Benchmark time:" outputs.
Expected Performance Results
See the reference results below for different system configurations with dual-socket CPU and NVIDIA Tesla boards for 1 to 4 nodes connected with 4xEDR InfiniBand.


Recommended System Configurations
Hardware Configuration
PC
Parameter
Specs
CPU Architecture
x86
System Memory
16-32GB
CPUs
2 (8+ CORES, 2+ GHz)
GPU Model
GeForce GTX TITAN X
GPUs
2-4
Servers
Parameter
Specs
CPU Architecture
x86
System Memory
32 GB
CPUs/Nodes
2 (8+ cores, 2+ GHz)
Total# of Nodes
1 – 10,000
GPU Model
NVIDIA Tesla
® P100
GPUs/Nodes
2 – 4
Interconnect
InfiniBand
Software Configuration
Software stack
Parameter
Version
OS
CentOS 6.2
GPU Driver
352.68 or newer
CUDA Toolkit
7.5 or newer
Intel Compilers
2016.0.047