Numba is an open-source, just-in-time compiler for Python code that developers can use to accelerate numerical functions on both CPUs and GPUs using standard Python functions.
Numba
What is Numba?
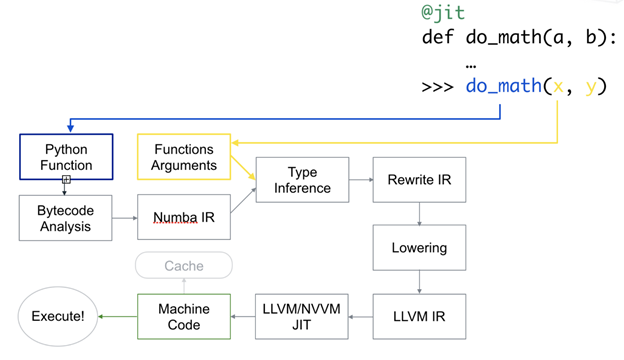
Numba translates Python byte-code to machine code immediately before execution to improve the execution speed.
Numba can be used to optimize CPU and GPU functions using callable Python objects called decorators. A decorator is a function that takes another function as input, modifies it, and returns the modified function to the user. This modularity reduces programming time and increases Python’s extensibility.
Numba also works with NumPy, an open-source Python library of complex mathematical operations designed for processing statistical data. When invoked with a decorator, Numa translates a subset of Python and/or NumPy code into bytecode automatically optimized for the environment. It uses LLVM, an open source API-oriented library for programmatically creating machine-native code. Numba offers several options for quickly parallelizing Python code for various CPU and GPU configurations, sometimes with only a single command. When used in conjunction with NumPy, Numba generates specialized code for different array data types and layouts to optimize performance.
Why Numba?
Python is a high-productivity dynamic programming language that’s widely used in data science. It’s extremely popular due to its clean and expressive syntax, standard data structures, comprehensive standard library, excellent documentation, broad ecosystem of libraries and tools, and large and open community. Perhaps most important, though, is the high productivity that a dynamically typed, interpreted language like Python enables.
But Python’s greatest strength can also be its greatest weakness. Its flexibility and typeless, high-level syntax can result in poor performance for data- and computation-intensive programs, as running native, compiled code is many times faster than running dynamic, interpreted code. For this reason, Python programmers concerned about efficiency often rewrite their innermost loops in C and call the compiled C functions from Python. There are a number of projects aimed at making this optimization easier, such as Cython, but they often require learning a new syntax. While Cython improves the performance significantly, it can require painstaking manual modification to Python code.
Numba was conceived as a much simpler alternative to Cython. One of its most appealing traits is that it doesn’t require learning a new syntax, replacing the Python interpreter, running a separate compilation step, or having a C/C++ compiler installed. Just applying the @jit Numba decorator to a Python function is enough. This enables compilation at run time (this is “Just-in-Time”, or JIT compilation). Numba’s ability to dynamically compile code means that you don’t give up the flexibility of Python. Also, numerical algorithms within Python programs that are compiled by Numba can approach the speeds of programs written in the compiled C or FORTRAN languages and run up to 100 times faster than the same procedures executed by the native Python interpreter. This is a huge step toward providing the ideal combination of high-productivity programming and high-performance computing.
Numba is designed for array-oriented computing tasks, much like the widely used NumPy library. The data parallelism in array-oriented computing tasks is a natural fit for accelerators like GPUs. Numba understands NumPy array types, and uses them to generate efficient compiled code for execution on GPUs or multicore CPUs. The programming effort required can be as simple as adding a @vectorize function decorator to instruct Numba to generate a compiled, vectorized version of the function at run time. This lets it be used to process arrays of data in parallel on the GPU.
In addition to JIT compiling NumPy array code for the CPU or GPU, Numba exposes “CUDA Python”: the NVIDIA® CUDA® programming model for NVIDIA GPUs in Python syntax. By speeding up Python, its ability is extended from a glue language to a complete programming environment that can execute numeric code efficiently.
The combination of Numba with other tools in the Python data science ecosystem transforms the experience of GPU computing. The Jupyter Notebook provides a browser-based document creation environment that allows the combination of Markdown text, executable code, and graphical output of plots and images. Jupyter has become very popular for teaching, documenting scientific analyses, and interactive prototyping.
Numba has been tested on more than 200 different platform configurations. It runs on Windows, Apple Macintosh, and Linux operating systems on top of Intel and AMD x86, POWER8/9, and ARM CPUs, as well as both NVIDIA and AMD GPUs. Precompiled binaries are available for most systems.
Use Cases
Scientific Computing
There are many applications of array processing, ranging from geographic information systems to calculating complex geometric shapes. Arrays are used by telecommunication companies to optimize the design of wireless networks and the healthcare researchers to analyze waveforms that have information about internal organs. Arrays can be used to reduce external noise in language processing, astronomical imaging, and radar/sonar.
Languages like Python have made applications in these areas available to developers without extensive mathematical training. However, Python’s performance shortcomings in numerically intensive calculations can severely impact processing speed in some applications. Numba is one of several solutions and is considered by many people to be the simplest to use, making it particularly valuable for students and developers without experience in more complex languages like C.
Why Numba Matters to Data Scientists
Iterative development is a useful time-saving device in data science because it enables developers to continually improve programs based upon observation of results. Interpreted languages like Python are particularly useful in this context. However, Python’s performance limitations in highly mathematical operations can create bottlenecks that slow overall processing speed and limit developer productivity.
Numba addresses this problem by providing developers with a simple way to call a compiler function that can dramatically improve performance over large calculations and arrays. Numba is easy to learn and saves data scientists from the often complex task of writing subroutines in compiled languages to optimize speed.
Why Numba Is Better on GPUs
Architecturally, the CPU is composed of just a few cores with lots of cache memory that can handle a few software threads at a time. In contrast, a GPU is composed of hundreds of cores that can handle thousands of threads simultaneously.

Numba supports CUDA GPU programming by directly compiling a restricted subset of Python code into CUDA kernels and device functions following the CUDA execution model. Kernels written in Numba appear to have direct access to NumPy arrays, which are transferred between the CPU and the GPU automatically. This gives Python developers an easy entry into GPU-accelerated computing and a path to learn how to apply increasingly sophisticated CUDA code without learning new syntax or languages. With CUDA Python and Numba, you get the best of both worlds: rapid iterative development with Python combined with the speed of a compiled language targeting both CPUs and NVIDIA GPUs.
One test using a server with an NVIDIA P100 GPU and an Intel Xeon E5-2698 v3 CPU found that CUDA Python Mandelbrot code compiled in Numba ran nearly 1,700 times faster than the pure Python version. The performance improvement was a result of several factors, including compilation, parallelization, and GPU acceleration compared to a single-threaded Python code on the CPU. However, it illustrates the acceleration that can be achieved with the addition of just a single GPU.
NVIDIA GPU-Accelerated, End-to-End Data Science
The NVIDIA RAPIDS™ suite of open-source software libraries, built on CUDA-X AI, provides the ability to execute end-to-end data science and analytics pipelines entirely on GPUs. It relies on NVIDIA CUDA primitives for low-level compute optimization, but exposes that GPU parallelism and high-bandwidth memory speed through user-friendly Python interfaces.
With the RAPIDS GPU DataFrame, data can be loaded onto GPUs using a Pandas-like interface, and then used for various connected machine learning and graph analytics algorithms without ever leaving the GPU. This level of interoperability is made possible through libraries like Apache Arrow, and allows acceleration for end-to-end pipelines—from data prep to machine learning to deep learning.

RAPIDS supports device memory sharing between many popular data science libraries. This keeps data on the GPU and avoids costly copying back and forth to host memory.

The RAPIDS team is developing, contributing, and collaborating closely with numerous open-source projects including Apache Arrow, Numba, XGBoost, Apache Spark, scikit-learn, and others to ensure that all the components of the GPU-accelerated data science ecosystem work smoothly together.